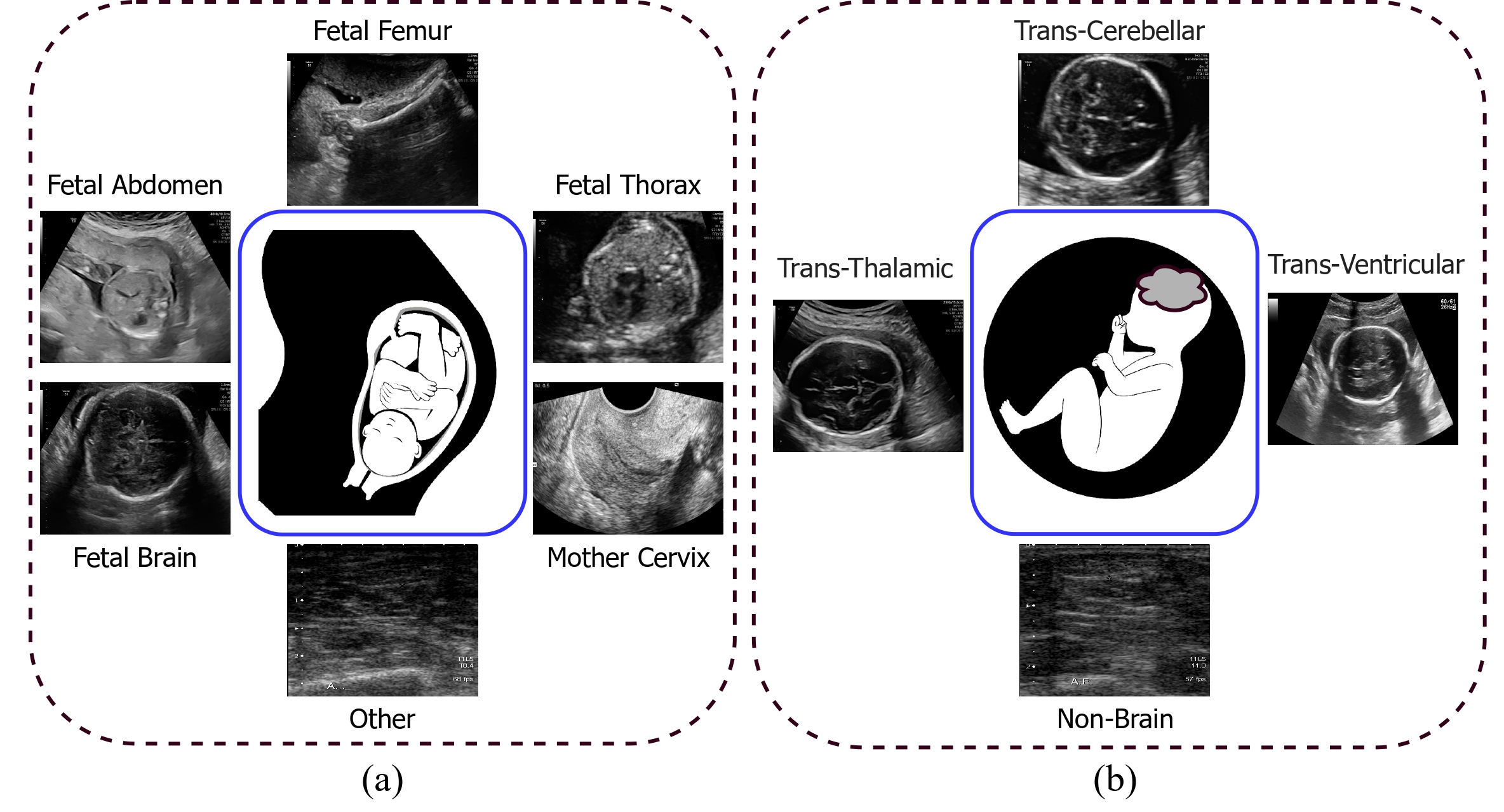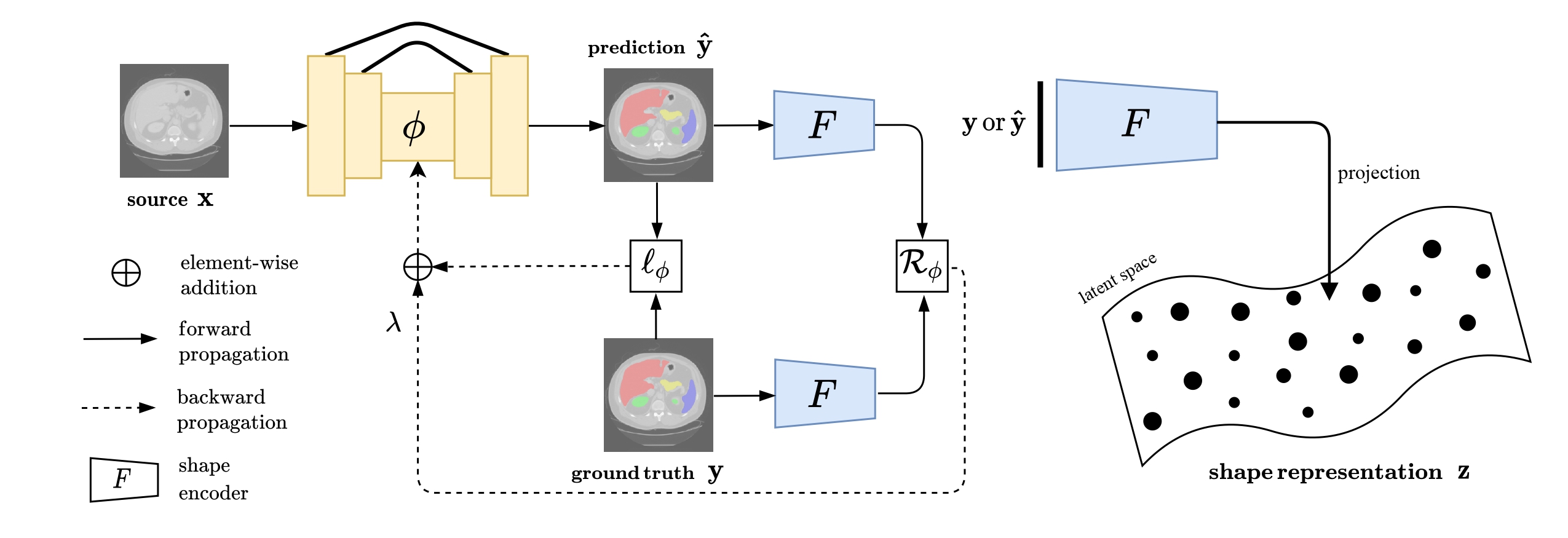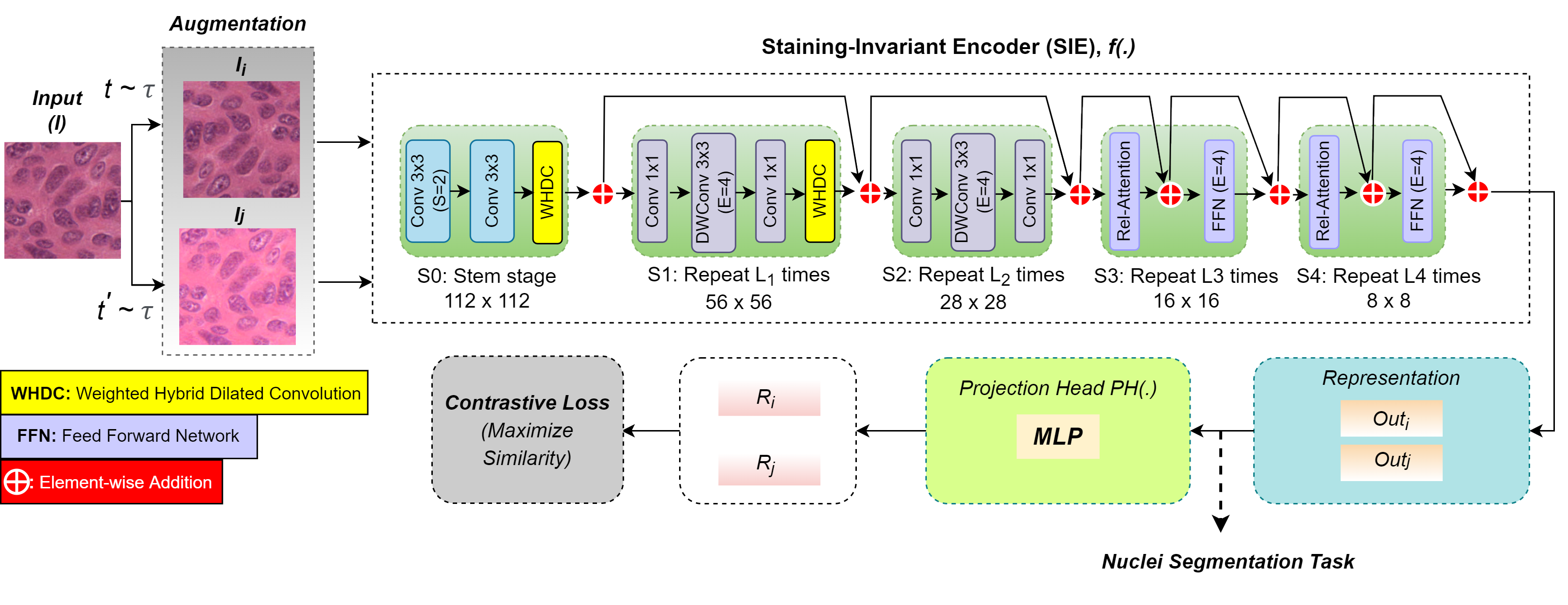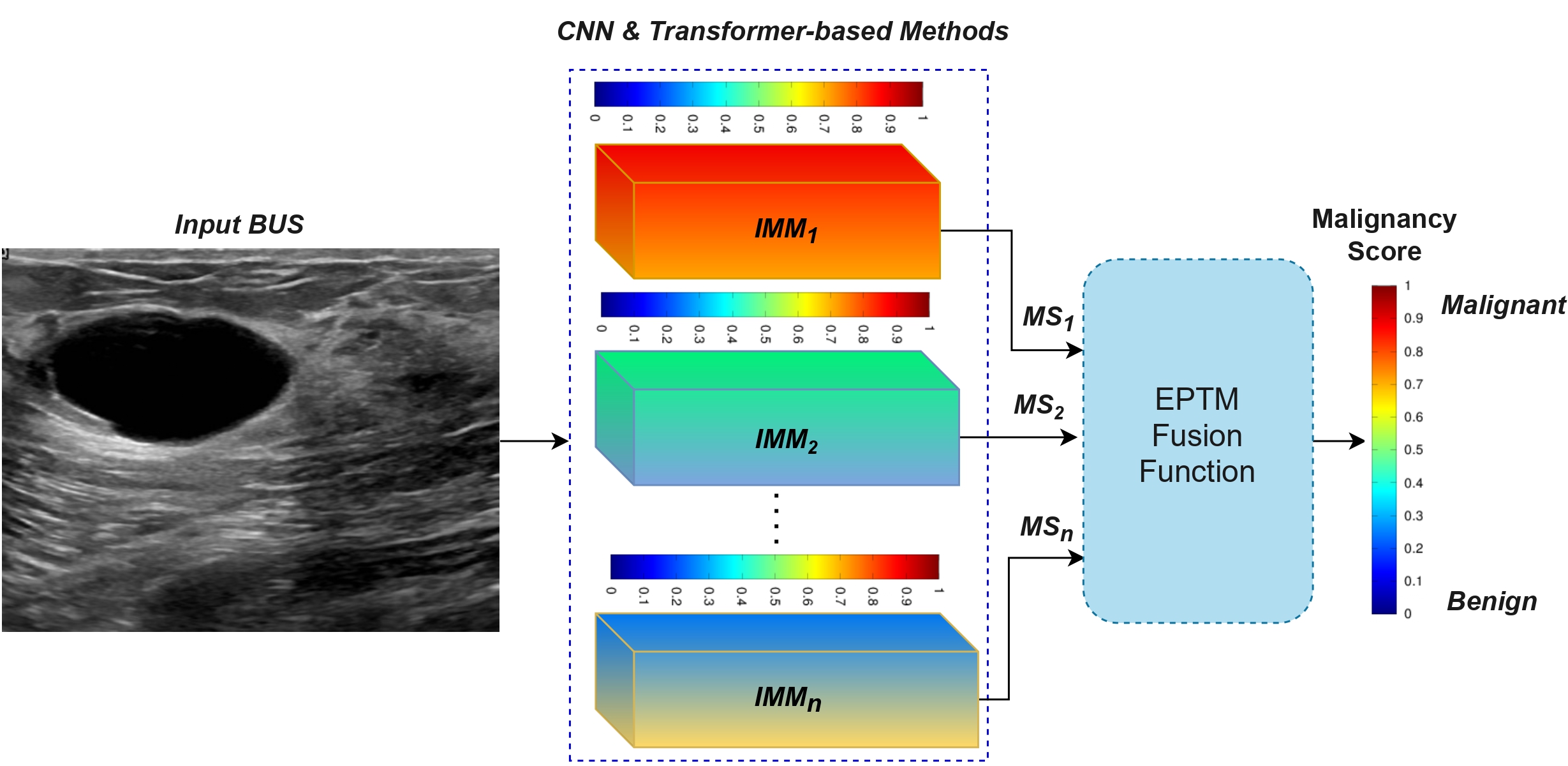
Vivek Kumar Singh, Ehab Mahmoud Mohamed, and Mohamed Abdel-Nasser. ” Aggregating efficient transformer and CNN networks using learnable fuzzy measure for breast tumor malignancy prediction in ultrasound images.” Neural Computing and Applications (2024): 1-17.
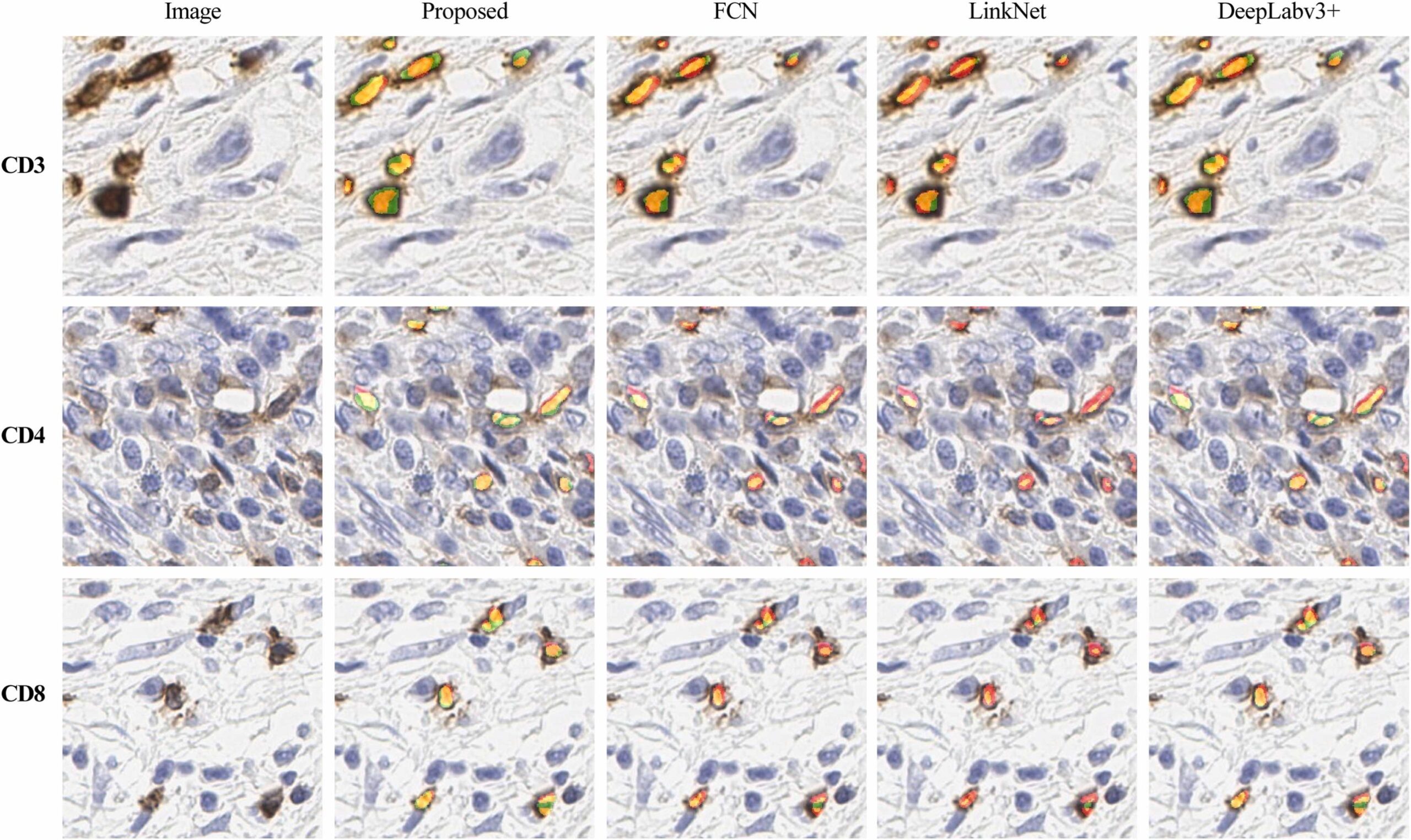
Makhlouf, Yasmine, Vivek Kumar Singh, Stephanie Craig, Aoife McArdle, Dominique French, Maurice B. Loughrey, Nicola Oliver et al.” True-T–Improving T-cell response quantification with holistic artificial intelligence-based prediction in immunohistochemistry images.” Computational and Structural Biotechnology Journal 23 (2024): 174-185.
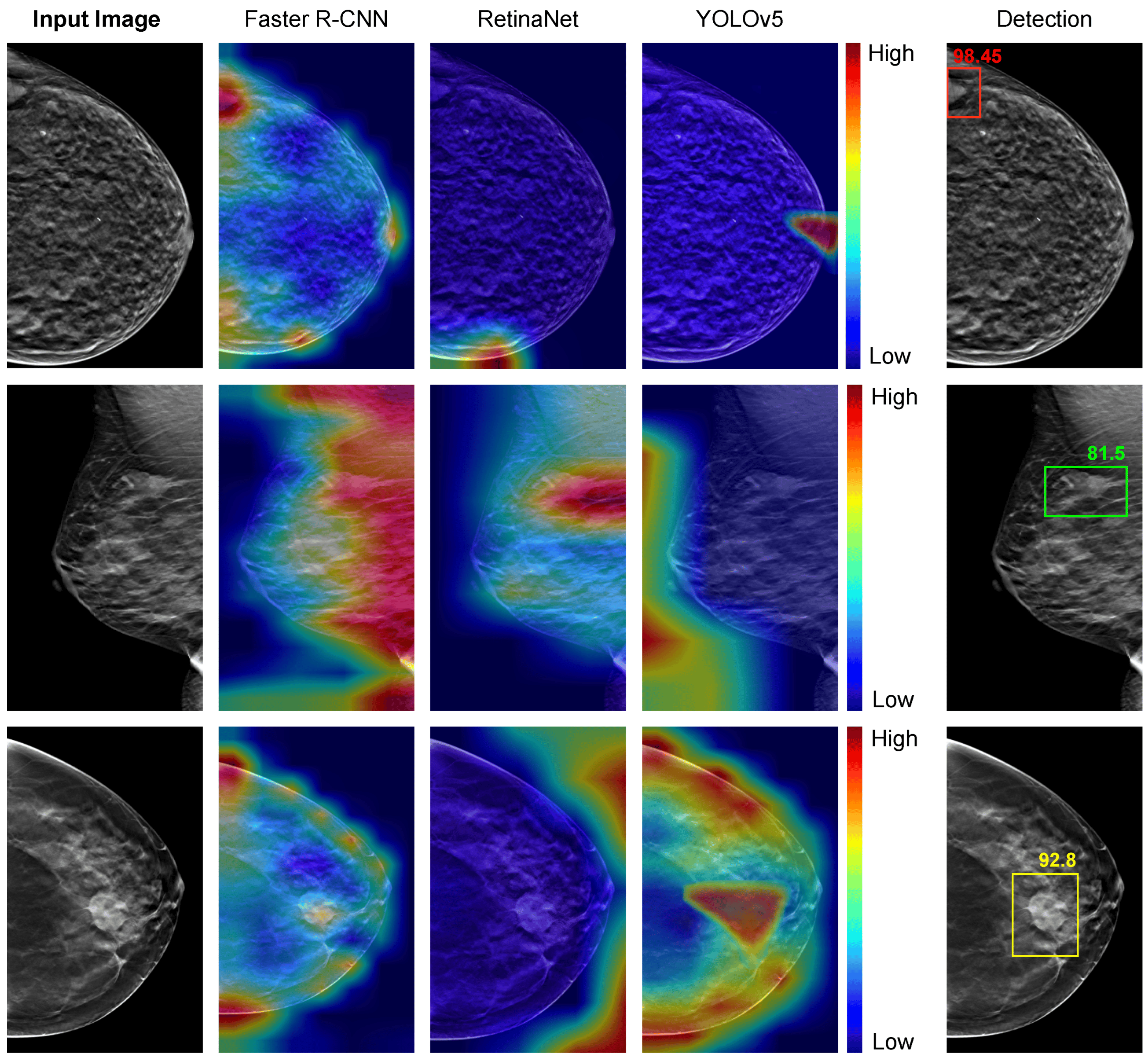
Hassan, Loay, Adel Saleh, Vivek Kumar Singh, Domenec Puig, and Mohamed Abdel-Nasser. ”Detecting Breast Tumors in Tomosynthesis Images Utilizing Deep Learning-Based Dynamic Ensemble Approach.” Computers 12, no. 11: 220.